Numeric posterior of the independent variable
In this notebook, we demonstrate the use of infer_independent and predict_independent. Given observations \(Y_{obs}\) made at the same corresponding independent variable (e.g. several slightly different measurements of the same glucose concentration), infer_independent will return a probability distribution taking all observations into account. predict_independent will only return the median of a distribution under the assumption that this was the only observation. For a uniform prior, the posterior inferred by MCMC yields the same result as a numerically integrated likelihood function, which is demonstrated at the end of the notebook.
[1]:
import ipywidgets
from matplotlib import pyplot
import numpy
import pymc as pm
import scipy.optimize
import scipy.stats
import calibr8
Synthetic groundtruth
[2]:
θ_true = (0.1, 4, 15, 0.3, 1)
σ_true = 0.05
df_true = 1.5
X = numpy.linspace(0.1, 30, 24)
X_dense = numpy.linspace(0, 30, 100)
Y = calibr8.asymmetric_logistic(X, θ_true)
numpy.random.seed(230620)
Y = scipy.stats.t.rvs(loc=Y, scale=σ_true, df=df_true)
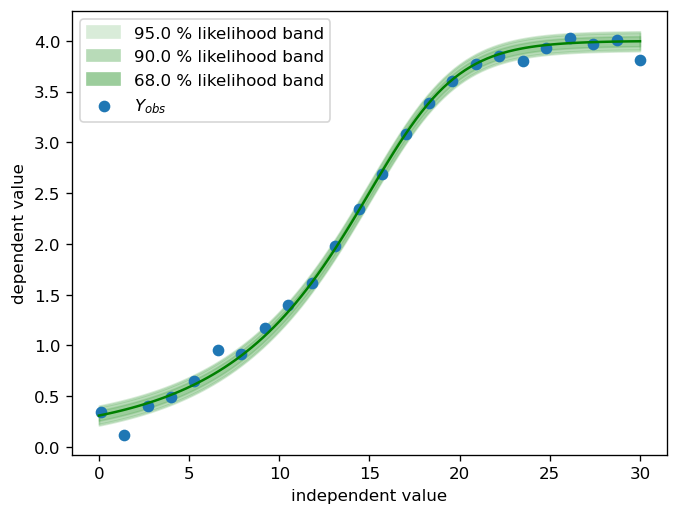
fig, ax = pyplot.subplots(dpi=120)
calibr8.plot_norm_band(ax, X_dense, calibr8.asymmetric_logistic(X_dense, θ_true), σ_true)
ax.scatter(X, Y, label='$Y_{obs}$')
ax.set_xlabel('independent value')
ax.set_ylabel('dependent value')
ax.legend()
pyplot.show()
Model definition
[3]:
class AsymmetricTModelV1(calibr8.BaseAsymmetricLogisticT):
def __init__(self):
super().__init__(independent_key='independent', dependent_key='dependent', scale_degree=0)
Model fit
[4]:
model = AsymmetricTModelV1()
calibr8.fit_scipy(
model,
independent=X, dependent=Y,
theta_guess=[
0, # lower limit approximately at 0 ?
4.5, # upper limit maybe around 4.5 ?
12, # inflection point maybe at arond 12 ?
1/5, # approximate slope at inflection point
2, # asymmetry paramter: the inflection point is near the upper limit -> positive value
] + [0.01, 5],
theta_bounds=[
(-numpy.inf, 1), # L_L
(3.5, numpy.inf), # L_U
(10, 20), # I_x
(1/20, 1), # S
(-3, 3), # c
] + [(0.01, 0.5), (0.5, 50)]
)
fig, axs = calibr8.plot_model(model)
fig.tight_layout()
pyplot.show()
print(f'The following estimations were made for {model.theta_names}: {model.theta_fitted}')
C:\Users\osthege\AppData\Local\Temp\ipykernel_356\3130538705.py:22: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
fig.tight_layout()
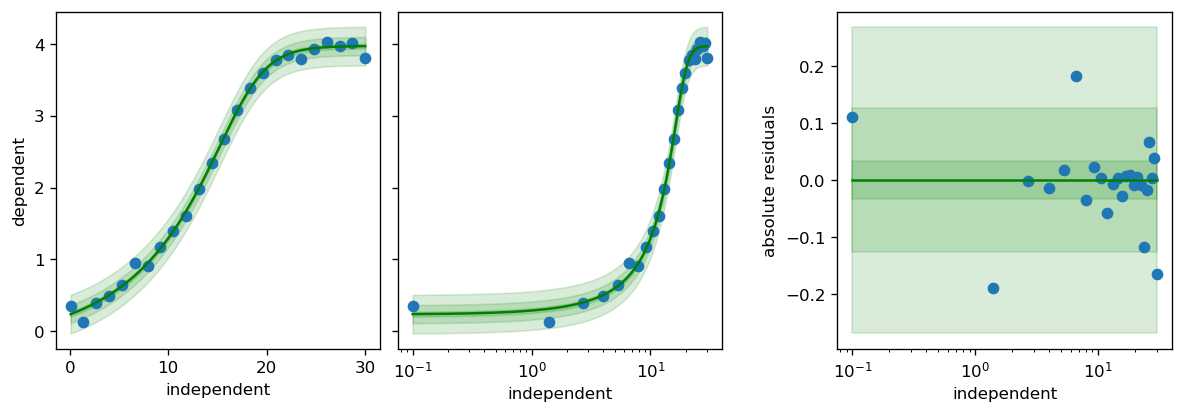
The following estimations were made for ('L_L', 'L_U', 'I_x', 'S', 'c', 'scale_0', 'df'): (-0.1618649667081871, 3.974967996155461, 15.262602665464405, 0.2865868328729957, 1.3554398193364028, 0.017449300261076536, 0.9251045856162367)
Visualization
[5]:
def plot(y_obs=0.5, a=0, b=30, steps=1000, ci_prob=0.95):
y_obs = numpy.atleast_1d(y_obs)
fig, ax = pyplot.subplots(dpi=120)
posterior = model.infer_independent(y_obs, lower=a, upper=b, steps=steps, ci_prob=ci_prob)
ax.plot(posterior.hdi_x, posterior.hdi_pdf, color='b')
# Plot the individual predictions from predict_independent for comparison
for yo in y_obs:
ax.axvline(model.predict_independent(yo), linestyle='--', color='b', alpha=0.3)
ax.plot([], linestyle='--', label='predict_independent', color='b', alpha=0.3) #to get only one legend
# For multiple observations of the same independent variable,
# the median of the posterior will differ from the two predict_independent results
ax.annotate('median', xy=(posterior.median, 0),
xytext=(posterior.median, numpy.max(posterior.hdi_pdf)*0.2), xycoords='data',
arrowprops=dict(facecolor='b', edgecolor='b', shrink=0.05),
horizontalalignment='right', verticalalignment='top',
ha='center'
)
ax.legend()
ax.set_xlabel('independent variable')
ax.set_ylabel(r'$p(x\ |\ \vec{y}_{obs})$')
ax.set_ylim(0)
return fig, ax
Plot for one observation, where predict_independent is similar to the median
[6]:
def iplot(y_obs=0.5, a=0, b=30, steps=1000, ci_prob=0.95):
plot(y_obs, a, b, steps, ci_prob)
pyplot.show()
ipywidgets.interact(
iplot,
y_obs=θ_true[0:2],
a=ipywidgets.fixed(0),
b=ipywidgets.fixed(30),
steps=ipywidgets.fixed(1000),
percentiles=ipywidgets.fixed((0.025, 0.975))
);
Comparison with MCMC for two observations
[7]:
y_obs = numpy.array([3.85,3.9])
a = 0
b = 30
theta = model.theta_fitted
with pm.Model():
prior = pm.Uniform(model.independent_key, lower=a, upper=b, shape=(1,))
mu, scale, df = model.predict_dependent(prior, theta=theta)
pm.StudentT('likelihood', nu=df, mu=mu, sigma=scale, observed=y_obs, shape=(1,))
idata = pm.sample(10_000, cores=1, return_inferencedata=True)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Sequential sampling (2 chains in 1 job)
NUTS: [independent]
100.00% [11000/11000 00:08<00:00 Sampling chain 0, 0 divergences]
100.00% [11000/11000 00:07<00:00 Sampling chain 1, 0 divergences]
Sampling 2 chains for 1_000 tune and 10_000 draw iterations (2_000 + 20_000 draws total) took 16 seconds.
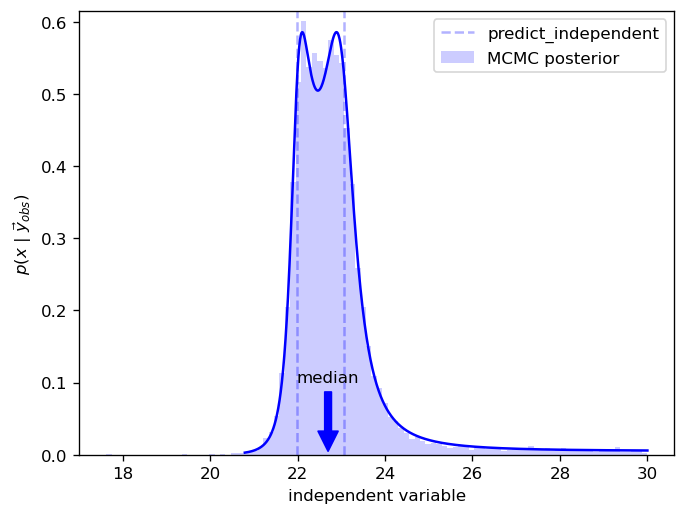
Here we can see how the median differs from predict_independent results
[8]:
fig, ax = plot(y_obs=y_obs, ci_prob=0.999)
ax.hist(
idata.posterior['independent'].stack(sample=('chain', 'draw')).values.T,
density=True, bins=100, color="b", alpha=0.2,
label='MCMC posterior'
)
ax.legend()
pyplot.show()
[9]:
%load_ext watermark
%watermark -n -u -v -iv -w
Last updated: Tue Dec 13 2022
Python implementation: CPython
Python version : 3.8.13
IPython version : 8.6.0
sys : 3.8.13 | packaged by conda-forge | (default, Mar 25 2022, 05:59:45) [MSC v.1929 64 bit (AMD64)]
ipywidgets: 8.0.2
scipy : 1.9.3
pymc : 4.4.0
calibr8 : 7.0.0
numpy : 1.23.4
matplotlib: 3.6.2
Watermark: 2.3.1
[ ]: